App is alternatively spliced in a model of neurodegeneration: implications for synaptic pathology in the pathogenesis of neurologic disease
Kaitland Fior
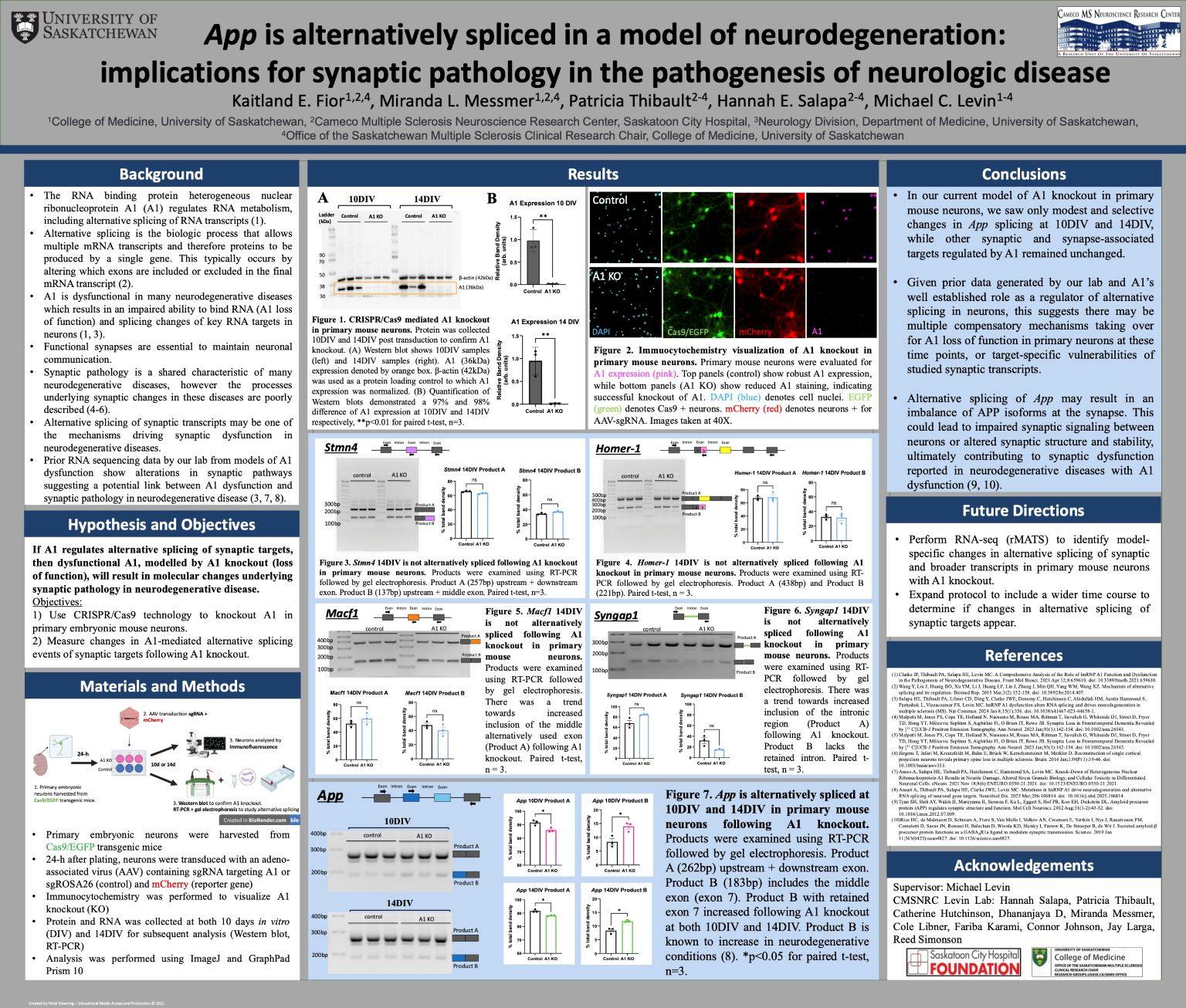
Background: Dysfunction of the RNA binding protein heterogeneous nuclear ribonucleoprotein A1 (A1) occurs in neurodegenerative diseases, resulting in loss of RNA binding, alternative RNA splicing, and changes in synaptic biology. Both splicing changes and synapse dysfunction are central to neurodegenerative disease. We therefore hypothesize that A1 loss of function alters splicing of synaptic targets, a potential mechanism driving synaptic dysfunction in neurodegenerative disease.
Methods: Cultured primary neurons from Cas9/EGFP transgenic mice were transduced with viruses containing guide RNA targeting A1 (A1 knockout) and Rosa26 (control). Protein and RNA were collected at 10 and 14 days in vitro (DIV). Immunocytochemistry and western blotting were used to confirm A1 knockout. RT-PCR and DNA gel electrophoresis were used to investigate splicing of synapse-related RNA targets.
Results: Knockout was confirmed with >97% decrease in A1 expression compared to control neurons (p<0.01, paired t-test). Alternative splicing analysis showed no significant difference of synaptic targets, Stmn4, Homer-1, and Syngap1 following A1 knockout. Importantly, there was a change in App splicing at 10DIV and 14DIV (p<0.05, paired t-test).
Conclusion: Considering the critical role that App plays in synapse biology, these results suggest that loss of A1 function precipitates abnormal App alternative RNA splicing, which may contribute to synaptopathology in neurologic disease.
Methods: Cultured primary neurons from Cas9/EGFP transgenic mice were transduced with viruses containing guide RNA targeting A1 (A1 knockout) and Rosa26 (control). Protein and RNA were collected at 10 and 14 days in vitro (DIV). Immunocytochemistry and western blotting were used to confirm A1 knockout. RT-PCR and DNA gel electrophoresis were used to investigate splicing of synapse-related RNA targets.
Results: Knockout was confirmed with >97% decrease in A1 expression compared to control neurons (p<0.01, paired t-test). Alternative splicing analysis showed no significant difference of synaptic targets, Stmn4, Homer-1, and Syngap1 following A1 knockout. Importantly, there was a change in App splicing at 10DIV and 14DIV (p<0.05, paired t-test).
Conclusion: Considering the critical role that App plays in synapse biology, these results suggest that loss of A1 function precipitates abnormal App alternative RNA splicing, which may contribute to synaptopathology in neurologic disease.
