
Dr. Joyce Wilson BSc, MSc, PhD
Professor Biochemistry, Microbiology & Immunology- Address
- 6B67, Health Sciences
Research Area(s)
- HCV interaction with the host miRNA machinery
About
Currently accepting applications for graduate students.
Research Overview:
The current research in the Wilson lab centers on study of SARS-CoV-2, the virus that causes COVID-19, and flaviviruses that usurp microRNAs to augment their lifecycles.
SARS-CoV-2 and the COVID-19 pandemic
The response to the COVID-19 pandemic is a remarkable testament to scientific achievement. In under a year, society went from vulnerability to the SARS-CoV-2 virus to the establishment of protection through vaccines. These vaccines, combined with widespread immunization campaigns, allowed the world to cautiously resume normalcy. However, it's important to acknowledge that the pandemic is far from over.
Persistent waves of SARS-CoV-2 infections across the global population continue to exert pressure and give rise to new virus variants. In this ever-evolving landscape, the need for innovative treatments and an understanding of the pathogenic potential of emerging variants remains imperative. This is not only essential for our ongoing battle against COVID-19 but also for our preparedness in confronting potential future pandemics.
Project 1: Unveiling Host Dependence Factors and Drug Repurposing Strategies via CRISPR Knockout Screens in SARS-CoV-2 Infections’
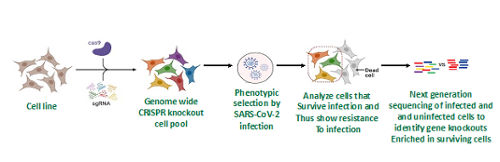
The goal of the research team, Dr. Joyce Wilson, Franco Vizeacoumar, and Darryl Falzarano is to identify existing therapeutics that can be re-purposed to treat SARS-CoV2 infections. During an infection, viruses must hijack host machinery to reproduce, and some of the machinery used by viruses are also involved in other human diseases, such as cancer. Thus, it is likely that therapeutics designed to treat other diseases and cancer also inhibit SARS-CoV2. However, the specific host factors used by SARS-CoV2 remain unknown and potential inhibitors undiscovered. Our strategy is to identify the host factors the virus needs to grow, and then test drugs that target them to see if they inhibit SARS-CoV-2. Thus far we have used a CRISPR knockout screen (Figure 1) to identify and confirmed 6 novel host factors that are used by SARS-CoV-2 and 3 drugs that target host factors that inhibit the virus.
-
Figure 1. Schematic diagram of the CRISPR-Cas9 knockout screening method to identify host dependency factors. A cell line that is highly susceptible to SARS-CoV-2 is transduced with Cas9 and a human genome wide gRNA library. The cells are then infected with SARS-CoV-2 and surviving cells grown and harvested. Surviving cells are hypothesized to have an enrichment of knockouts to genes required for SARS-CoV-2 infection and cell killing. gRNAs in surviving cells are identified by PCR and deep sequencing and their numbers compared with gRNA transduced cells that were not selected by virus infection. Enriched gRNAs target genes that are potential SARS-CoV-2 host dependency factors. Drugs targeting the potential host dependency factors were screened and 3 were found to inhibit SARS-CoV-2 in cultured cells. In addition, 6 potential dependency factors were validated by other methods. These drugs and host factors have the potential to be used in COVID-19 treatment strategies.
Impact: The inhibitors we discover are potential treatments for COVID-19 but might also be effective against other coronaviruses and thus help society prepare for future unforeseen coronavirus outbreaks.
Project 2: Advancing SARS-CoV-2 Replication Tools to Accelerate Drug Development
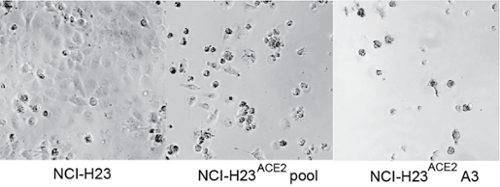
Our secondary objective is to generate innovative replication tools tailored for application in drug development. Notably, we have engineered a novel human lung cell line with susceptibility to SARS-CoV-2 (Figure 2). This cell line, rooted in the NCI-H23 human lung carcinoma cell line, has been rendered susceptible to SARS-CoV-2 through ACE2 receptor expression via transduction. Impressively, SARS-CoV-2 infects nearly 100% of these cells resulting in almost complete cell mortality, a crucial attribute for screens reliant on cell death for selection purposes.
-
Figure 2. The NCI-H23-Ace2 cell line transduced with ACE2, supports virus infection and showing high virus-induced CPE. Micrograph image of CPE of the labelled cell lines 24 hours post-infection with SARS-CoV-2 VIDO-01.
Additionally, we've constructed reporter variants of the SARS-CoV-2 ancestral strain (Wuhan-1), Delta, and Omicron (Figure 3). These engineered viruses incorporate the Nano Luciferase (Nluc) gene, and NLuc expression parallels virus replication levels. These reporter viruses serve as invaluable tools for drug screening endeavors.
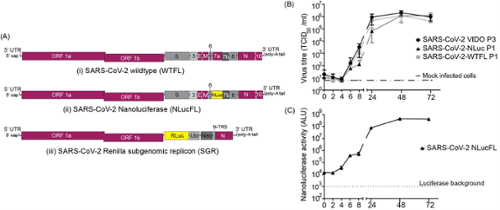
-
Figure 3. SARS-CoV-2 molecular clones and rescued viruses (A) The synthetic molecular clones for wild type full-length virus (WTFL) (i), NLuc expressing reporter virus (NLucFL) (ii), and sub-genomic replicon (SGR) (iii) are represented schematically. In the clone BAC plasmid, the genome is preceded by a T7 polymerase and a poly A sequence at the 3′ end. In NLucFL, the NLuc gene replaces ORF7a, (B) Replication kinetics of rescued SARS-CoV-2 WTFL and NLucFL were compared to SARS-CoV-2 wild type lab-isolated strain VIDO-01. (C) NLuc expression kinetics were observed to be robust, with peak values observed on Day 2 post-infection. The data are an average of three independent experiments, and error bars represent the standard deviation. ALU: arbitrary luminescence units.
Lastly, we've created a SARS-CoV-2 subgenomic replicon system. This system enables measurement of viral genome replication by assessing Nluc expression but does not generate infectious virus particles. Noteworthy advantages of this system include its facilitation of virus replication studies and antiviral drug screens, coupled with its cost-effectiveness and suitability for use in laboratories with lower containment requirements, reducing overall operational expenses.
Impact: The tools we have developed play a pivotal role in advancing the worldwide endeavor to create potent antiviral treatments for COVID-19.
Project 3: Decoding SARS-CoV-2 Variants: Unraveling the Impact of Mutations and Accessory Genes on Pathogenesis to Enhance Pandemic Preparedness
Waves of SARS-CoV-2 infections continue to spread through world human population and could pressure the emergence of new and potentially dangerous viruses. Global surveillance systems proficiently identify viruses with genetic changes, but the key challenge is distinguishing important genome alterations from benign ones. While there have been some advancement in. prediction of important mutations based on sequence information, and details regarding how specific spike mutations affect infections, the contributions of mutations in outside of spike, in other structural genes, non-structural genes, and accessory genes remain poorly characterized.
To bridge this knowledge gap, we have embarked on a comprehensive exploration focusing on the Delta and Omicron variants. By using reverse genetics and recombinant SARS-CoV-2 viruses this study will scrutinize the impacts of specific mutations, mutant genes, and the accessory gene ORF8 on virus growth, immune evasion, and disease progression. By unravelling the intricacies at play, the project aims to identify pivotal mutations, provide information required to improve predictions, and insight into pressures driving virus evolution during a pandemic.
Impact: Insight into the impact of specific mutations on the replication and pathogenesis of SARS-CoV-2 variants will guide public health strategies, vaccine design, and treatment approaches. understanding within this initiative is not only a means to navigate the present pandemic but is also a foundation for future pandemic preparedness.
Flaviviruses that usurp microRNAs to augment their lifecycles
A central focus of my laboratory research for many years has been the study of virus-host interactions between HCV, and BVDV and the host cell microRNAs (miRNAs) pathway. Micro RNAs (miRNAs) are endogenously expressed small regulatory RNAs that bind to the 3’ untranslated regions of cellular mRNAs and regulate their translation and stability. There are over 1500 identified human cellular miRNAs that directly regulate about one third of all human genes and links between miRNA regulation and diseases, including cancer attests to their importance in general biology.
Project 4: Understanding how host microRNAs augment the replication of Hepatitis C Virus (HCV) and Bovine Viral Diarrhea Virus (BVDV).
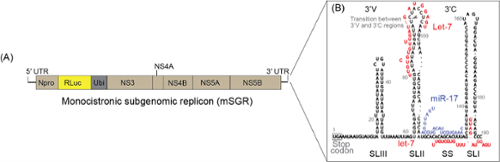
HCV and BVDV are uncommon pathogens that have ingeniously co-opted elements of the microRNA (miRNA) pathway to bolster their own replication processes. A liver-specific miRNA, known as miR-122, plays a pivotal role in facilitating HCV RNA accumulation during an infection, while BVDV co-opts miR-17 and let-7. These miRNAs engage in direct interactions with the viral genomes, miR-122 with the 5’ UTR of HCV (Figure 4), and miR-17 and let-7 with the 3’ UTR of BVDV (Figure 5) and emerging evidence suggests that these interactions serve to stabilize and finely modulate the structure and functionality of the viral genomes and induce early events in the initiation of assembly of replication complexes (Figure 4). This is a rather unconventional function for miRNAs, which typically function to repress translation and induce RNA degradation.
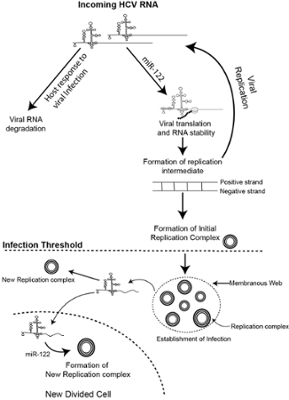
-
FIG. 4: Proposed model for the function of miR-122 in establishment of an HCV infection.
We propose that binding of miR-122 to the incoming viral RNA promotes viral translation and genome stability to allow the viral RNA to generate a threshold amount of viral protein required to initiate genome replication and the formation of virus replication complexes. After the establishment of replication complexes miR-122 may support ongoing virus replication by promoting the establishment of new replication complexes inside an infected cell or after an infected cell divides.
-
FIG. 5: Annealing pattern for miR-17 and let-7 on the BVDV 3’ UTRmiR-17 anneals to the single stranded (SS) region of the BVDV 5’ UTR and is required for efficient virus replication. let-7 has the capacity to anneal to the SS region and the top of stem loop II (SLII) but a functional role has only been established for let-7 annealing to SLII.
Both viruses intriguingly harness the same cellular machinery employed by host cells to promote their replication, a machinery primarily centered around the Ago2 protein. Our current research aims to uncover the exact mechanisms through which HCV and BVDV co-opt Ago2 and its associated proteins. Additionally, we are investigating whether these viruses exhibit a preference for or actively induce the production of specific phosphorylated isoforms of Ago2 to enhance their replication.
Impact: Our ongoing investigations delve into the intricate molecular mechanisms that underpin the subversion of cellular processes by HCV and BVDV, shedding light on the fascinating interplay between viruses and host cells.
For a list of publications please see Dr. Wilson’s Google Scholar profile:
https://scholar.google.com/citations?hl=en&user=U0C6JSIAAAAJ&view_op=list_works&sortby=pubdate
